Ribosomal Recoding
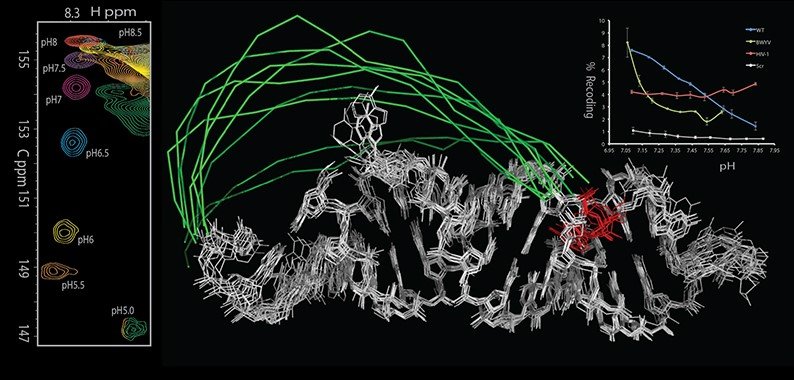
It has been well known that viruses employ ribosomal recoding — either by frameshifting or stop codon read-through — to regulate expression levels of various proteins. This not only ensures translation of proteins encoded in different frames but also yields very precise ratios of the proteins. We previously demonstrated (Houck-Loomis et al., Nature 2011) that the frequency of ribosomal readthrough in murine leukemia virus (MLV) is regulated by a dynamic mechanism, in which a recoding signal on the mRNA is maintained in a pre-existing, protonation-dependent equilibrium between two functional conformers: an active conformer that is permissive for recoding, and an alternate, inactive conformer that terminates translation. Importantly, the pKa of the equilibrium is such that at physiological pH the populations of the active and inactive form correlate with the critical levels of recoding and termination, respectively. We are currently working on determining recoding mechanisms in several other viral systems.
Additionally, recent research from other labs has identified many ribosomal recoding candidates in eukaryotic systems. As such, we are currently studying ribosomal readthrough systems in Drosophila and humans.